Use the Pluto CUT&RUN pipeline with a raw data upload
Streamlined analysis for your CUT&RUN experiment to identify DNA-protein interaction sites.
1. Create Experiment
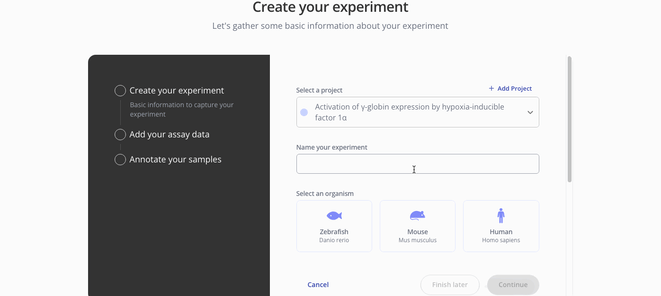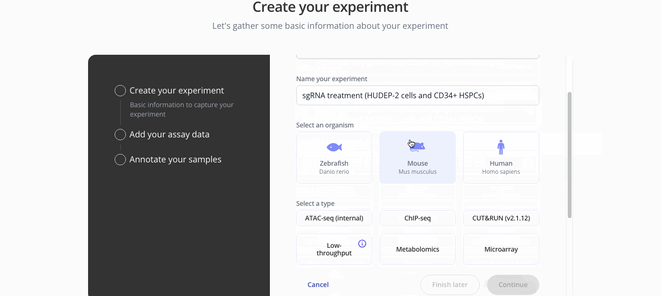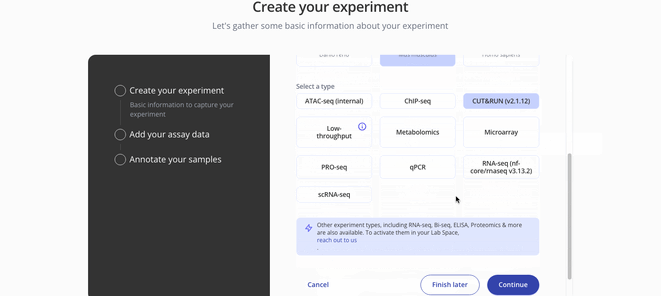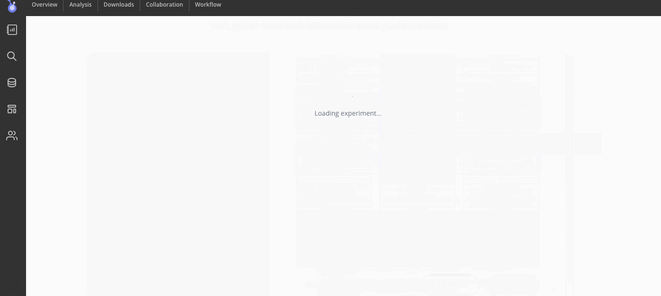
2. Upload assay data
First, you'll add your assay data as a FASTQ. For a FASTQ, your file when then undergo a series of preprocessing steps including the trimming of adapter sequences, alignment to the reference genome chosen when creating the experiment, filtering, peak calling and binding site identification.
Step 3. Add your sample annotations.
 Next, you will add your sample annotations with a .csv file containing the sample IDs from your assay data. We have a template available for download if needed. Here is an example of how to format your sample data.
Next, you will add your sample annotations with a .csv file containing the sample IDs from your assay data. We have a template available for download if needed. Here is an example of how to format your sample data. 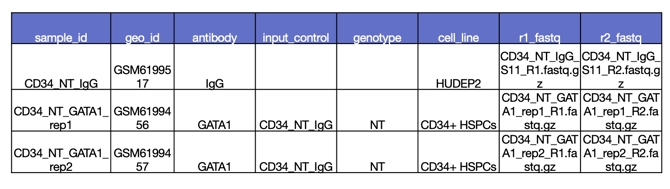
Step 4. Configure Pipeline

Your next step is to configure your pipeline. Here you'll enter information about the genome annotation you'd like to use, the library type and peak caller.
Genome - This field will populate with available genome annotations based on the organism you chose when creating the experiment. If this dataset is part of a larger experiment, make sure to select the same genome as your previous analyses in Pluto.
Library type - Paired end or single end. This is dependent on how your libraries were prepared for sequencing.
Peak Caller - On Pluto, you have the choice between MACS2 and SEACR for peak calling. Check out this article if you'd like some extra guidance on how to choose your peak caller. You'll also see some extra settings for peak type (narrow or broad) and peak p-value cutoff.
Step 5. Run your Analyses!
You'll be directed back to your experiment page where you can then start making plots and analyzing your data within our analysis tab. All analysis types for CUT&RUN are available with a FASTQ or raw data upload.
- Differential Binding (Heatmap and Volcano plots)
- IGV Peak plot
- Coverage (Tornado and Profile plots)
- Transcription Factor Enrichment analysis (Score Bar Plot)
- Gene Set Enrichment
- Summary Analysis (CPM normalized or raw values)
