Set up your single cell RNA-seq experiment in Pluto
Written by Caitlin Winkler, PhD
Single cell RNA sequencing (single cell RNA-seq or scRNA-seq) is a molecular sequencing technique that allows for investigation into cellular diversity and function. Unlike traditional bulk RNA-seq, single cell RNA-seq analyzes the transcriptional landscape within individual cells, uncovering intricate details about gene expression and cell-to-cell variability. By considering the transcriptome at a single cell resolution, scientists can being to unravel complex biological processes, uncover rare cell types, delineate developmental trajectories, and decode heterogeneity within tissues, offering new insights into the fundamental workings of living organisms.
Creating a single cell RNA-seq experiment in Pluto is easy, requires just a few clicks, and constitutes the initial Data upload phase of the Experiment roadmap.
Note: At this time, Pluto only supports 10x Genomics single cell RNA-seq data.
Step 1: Create experiment
To begin your analysis, navigate to your Lab Space and create a new Single-cell RNA sequencing experiment. Be sure to give your experiment an informative name, and pick the appropriate organism.
Step 2: Upload FASTQ files
Upload your FASTQ files directly to Pluto under the Add your assay data section of the Experiment wizard. If you are unsure how to upload your FASTQ files via the drag-and-drop uploader, or if you have a large batch of FASTQ files that you need to upload in bulk, contact the Pluto Support team to set up a direct transfer via your preferred cloud or other storage system. Learn more.
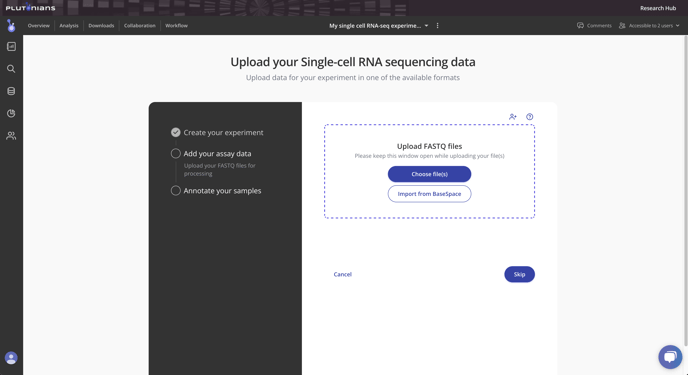
Tip: Does a sequencing company or another collaborator have your FASTQ files? No problem! You can invite them to the experiment as an Editor and they'll be able to upload the FASTQ files, or you can contact us about a direct file transfer from cloud storage or another location.
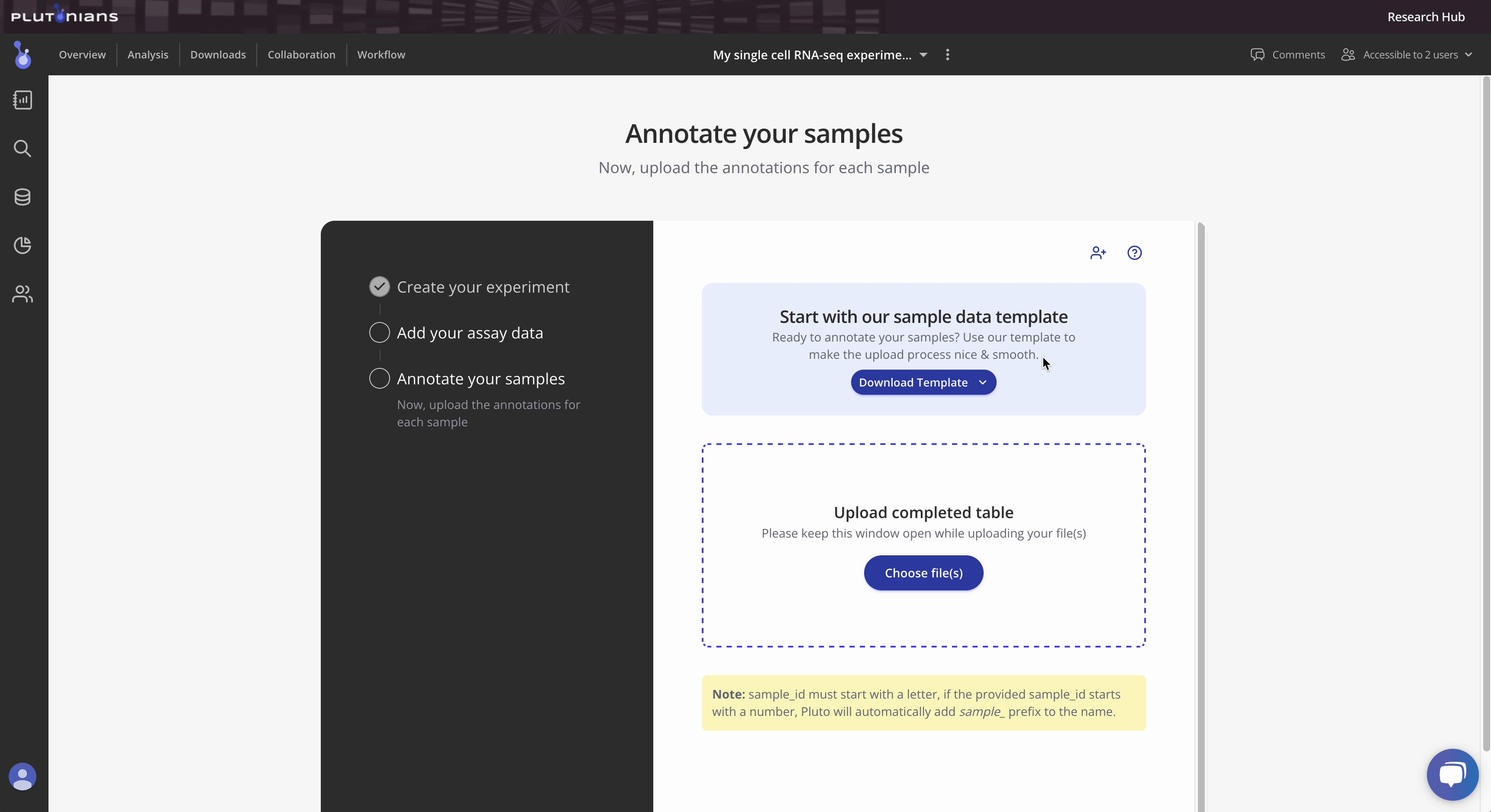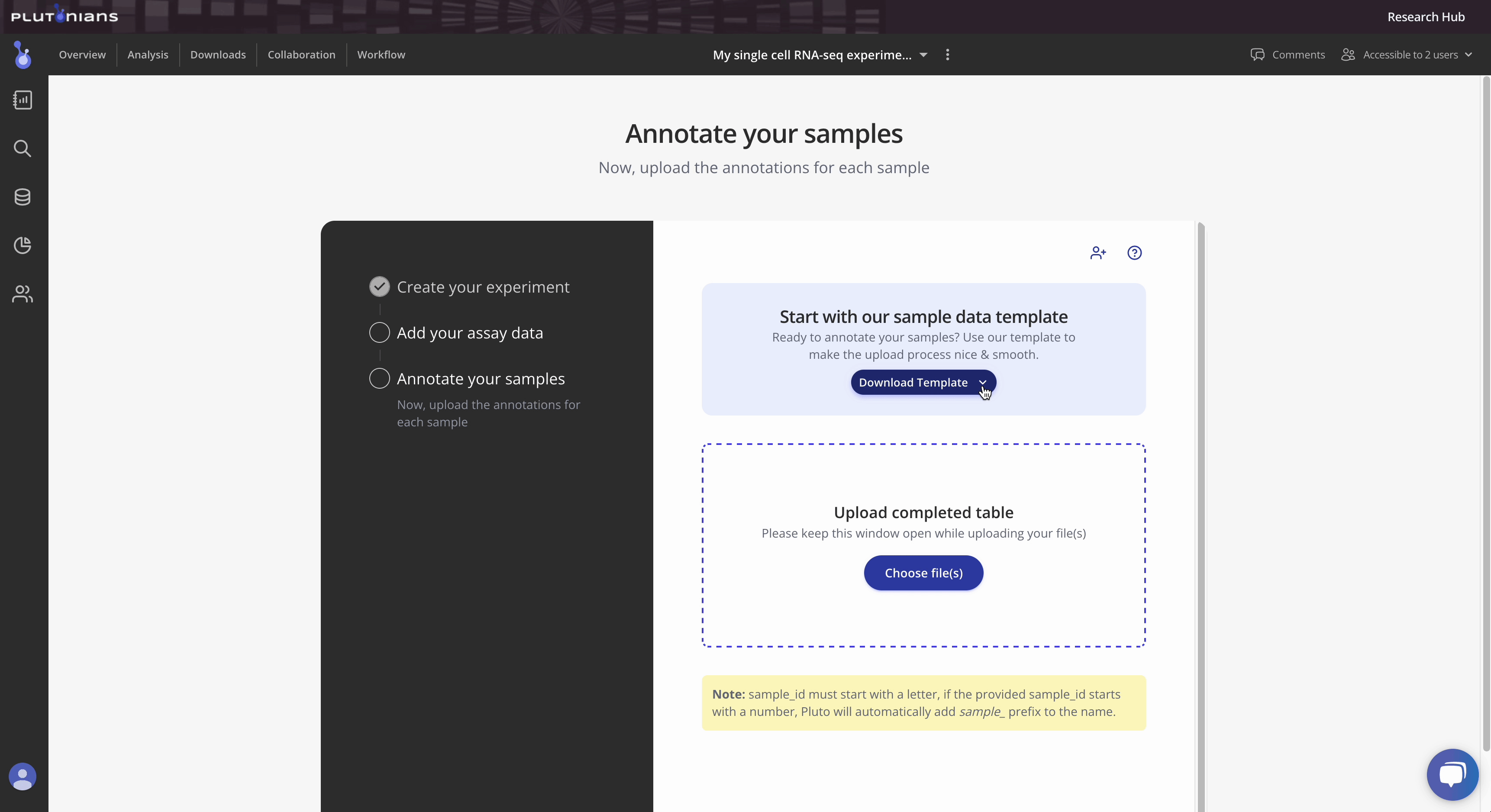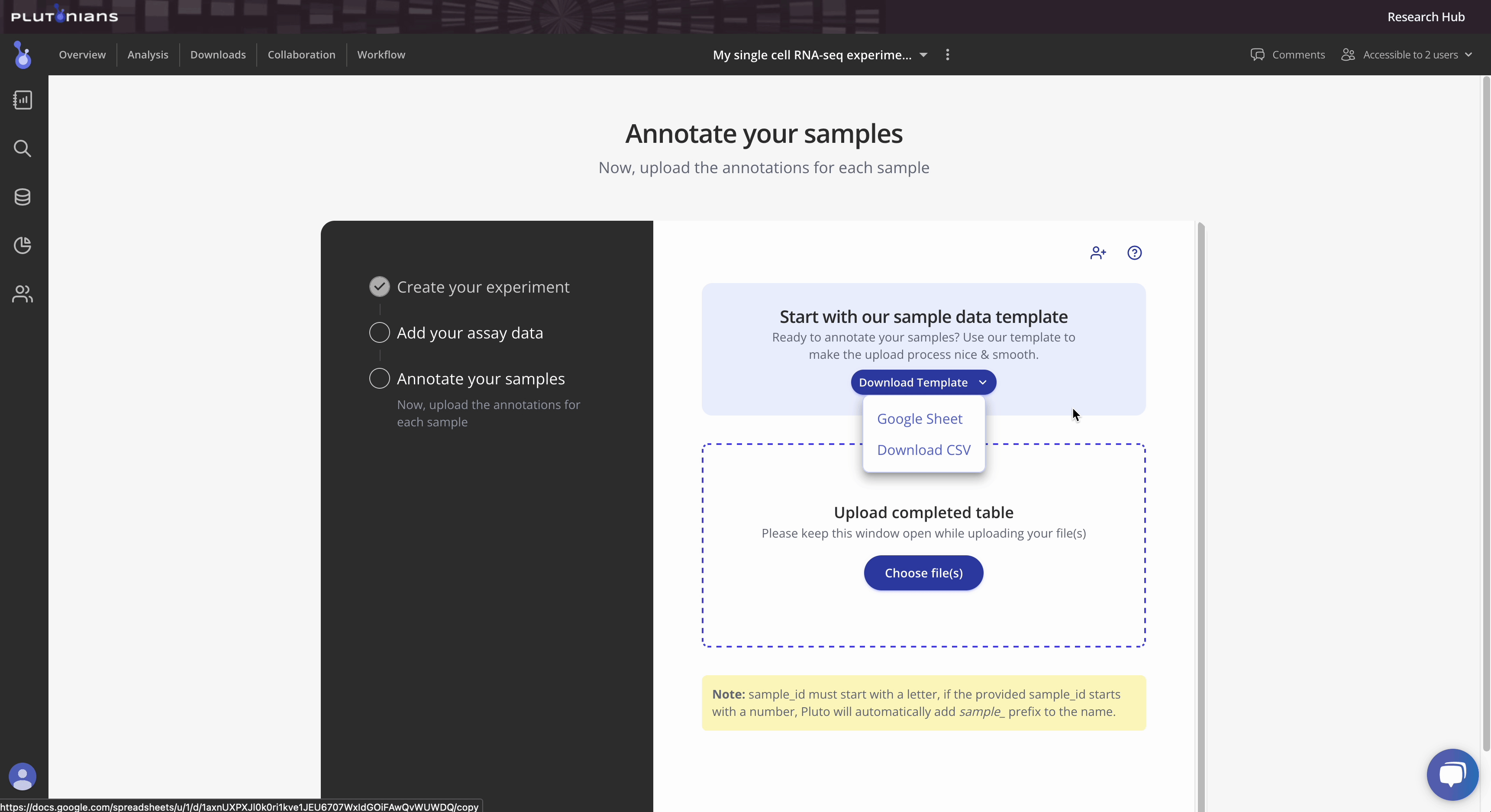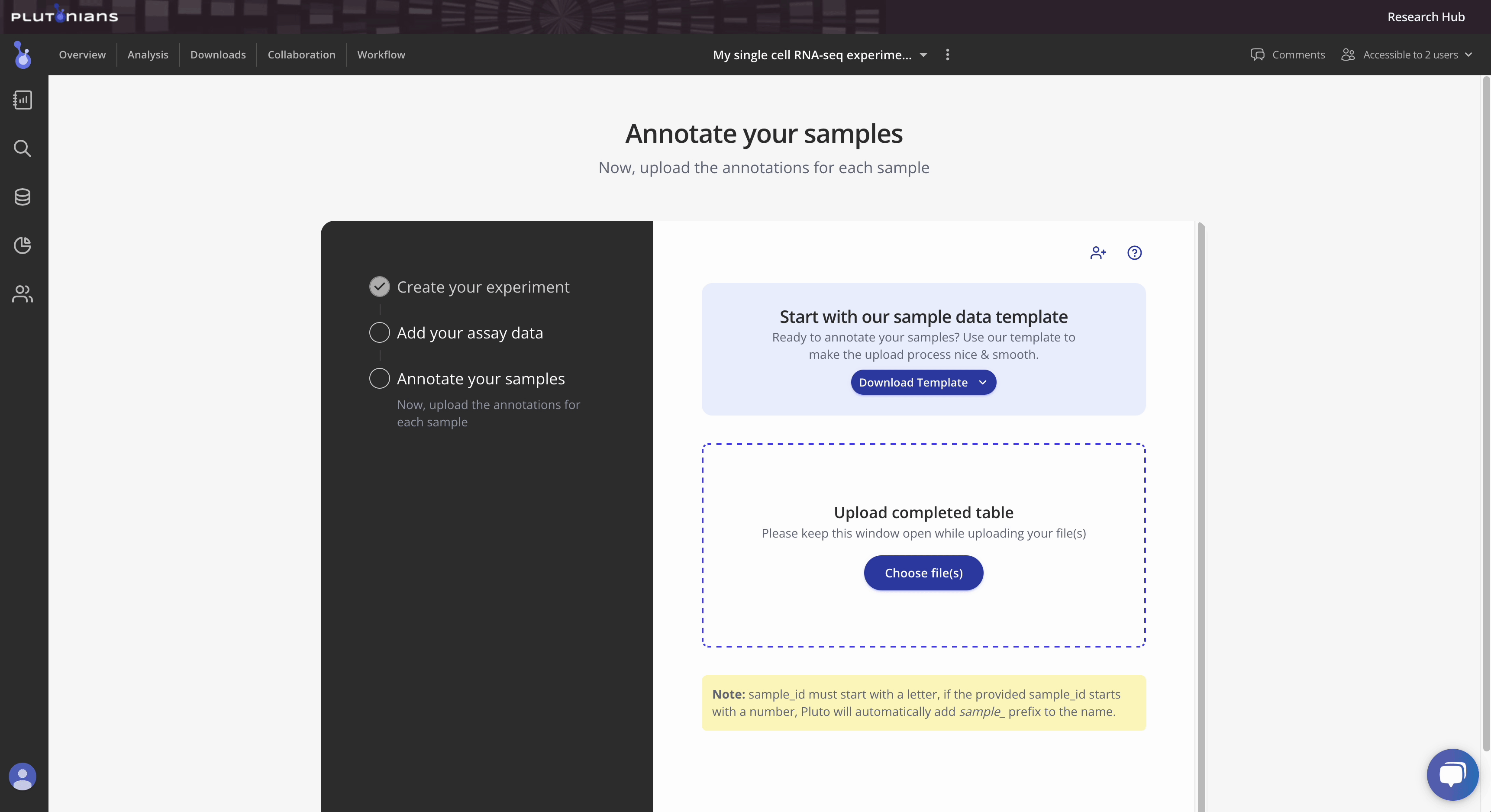
Step 3: Upload sample annotations
To define your samples, create a CSV file containing the FASTQ files from the data uploaded in Step 2 and their associated samples in a sample ID column, plus any additional columns for each relevant annotation or metadata you would like included in your analysis, such as age, sex, treatment, dose, or other conditions. Here's one example:
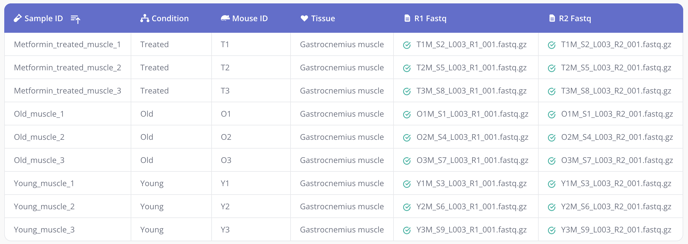
The sample ID column contains unique identifiers for your samples, so that each sample occupies one row in the table. The metadata columns can be used to group cells in different ways during analysis. Learn more.
Tip 1: Pluto sample tables can contain a variety of metadata. Take a look at our template here for more information. Additionally, you can access the template as either a Google sheet or downloadable CSV file from within the Experiment wizard.
Tip 2: Does your single cell experiment include samples that were sequenced across multiple lanes? No problem! FASTQ files that belong to samples with the same sample ID will be automatically merged by our single cell RNA-seq pipeline. Here's an example:
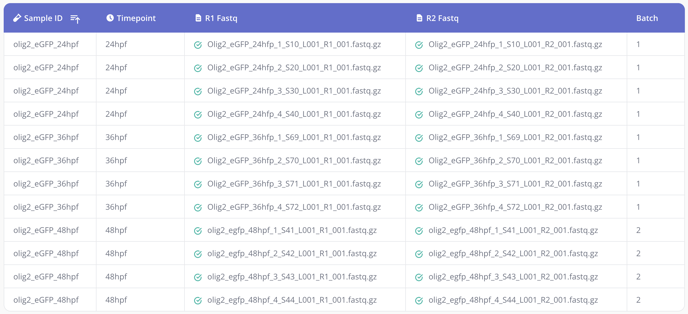
In this example, there are 3 samples - olig2_eGFP_24hpf, olig2_eGFP_36hpf, and olig2_eGFP_48hpf - that were sequenced across multiple lanes and will be merged by the pipeline.
Step 4: Kick off the single cell RNA-seq pipeline
To kick off the single cell RNA-seq pipeline (nf-core/scrnaseq) (1, 2), simply navigate to the Configure pipeline section of the Experiment wizard and select your desired reference genome and click the Run pipeline button. That's it! Once the pipeline has successfully completed, you will receive an email notification informing you that your data is ready to move on to the next phase in the Experiment roadmap: Preprocessing.
Here is an example workflow for creating a single cell RNA-seq experiment in Pluto from start to finish:
References
- Ewels et al. The nf-core framework for community-curated bioinformatics pipelines. Nature Biotechnology (2020). doi: 10.1038/s41587-020-0439-x
- nf-core/scrnaseq. doi: 10.5281/zenodo.3568187
