Pluto can import large data files from a variety of sources
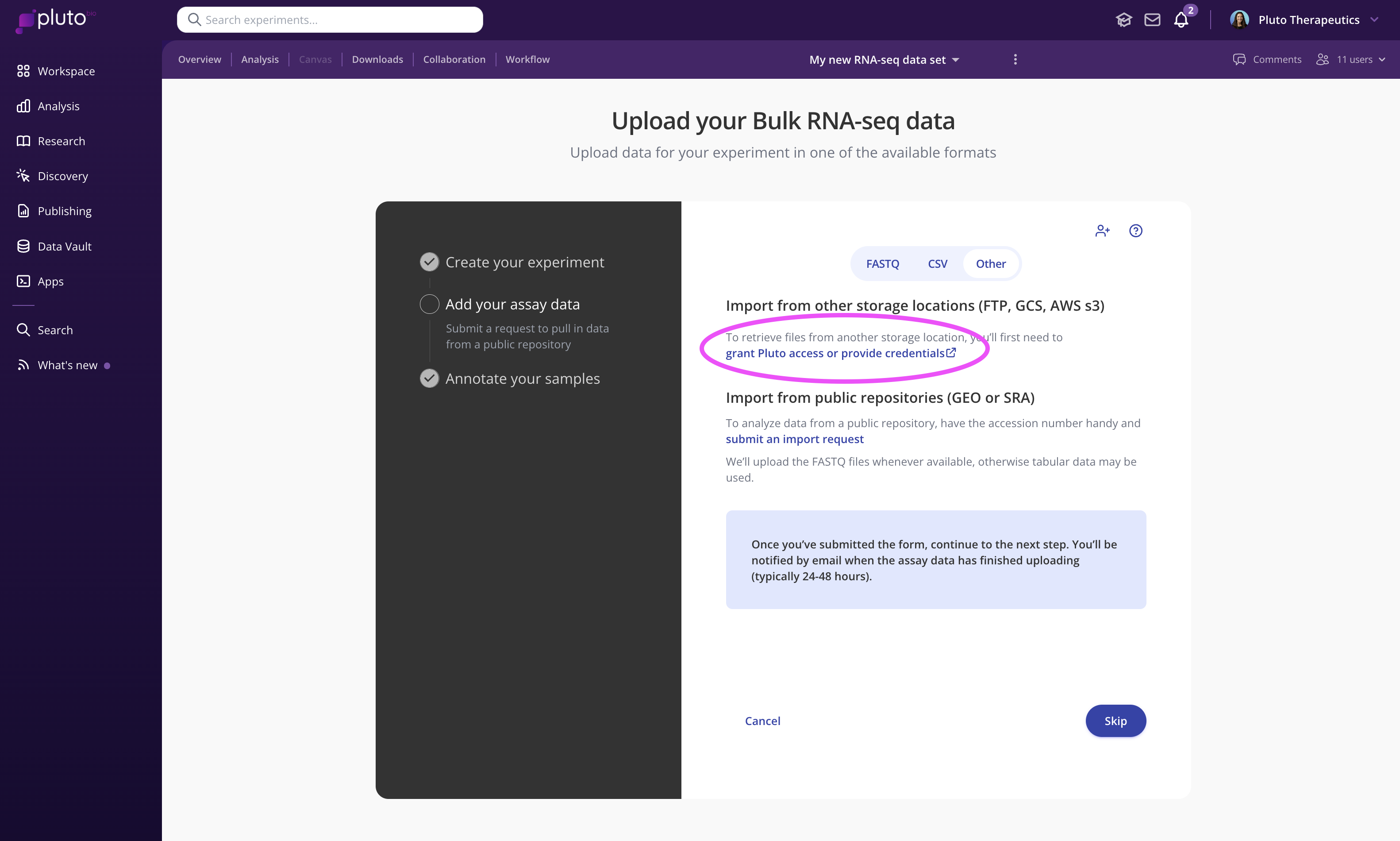
The first step is to create your experiment. During the "add your assay data" step, click "Other", and click the link under "Import from other storage locations (FTP, GCS, AWS s3)". This will direct you to a secure form for submitting the required information:
FTP
Large files stored with sequencing vendors and other third parties are often made available to download via FTP. In this case, you can share the FTP credentials you receive from a vendor to by submitting an FTP import form within the experiment creation process (see video above).
AWS and GCP
Globus
Create a shared Globus collection containing the files you wish to import, Share the collection with dataimport@pluto.bio, and submit the external storage import form within the experiment creation process (see video above).
Other cloud storage (Dropbox, Box, Google Drive, etc)
Create a shared folder containing your files. Share the folder with dataimport@pluto.bio, and submit the external storage import form within the experiment creation process (see video above).
BaseSpace
Import files directly from BaseSpace into your experiments in Pluto by enabling the BaseSpace integration in your Lab Space.
Once enabled, all members of your organization will see the "Import from BaseSpace" option during the FASTQ file upload step on any sequencing-based experiment type:
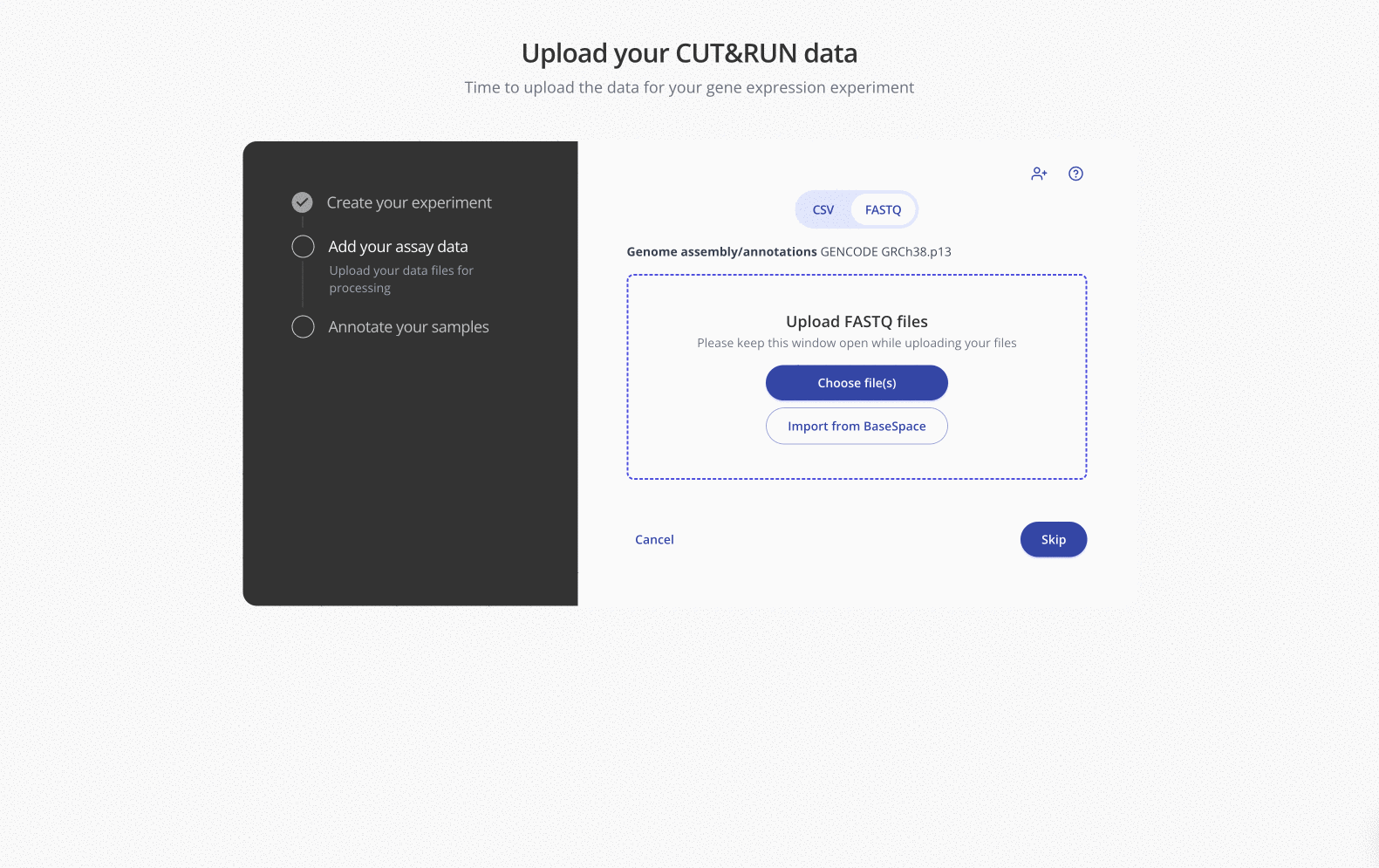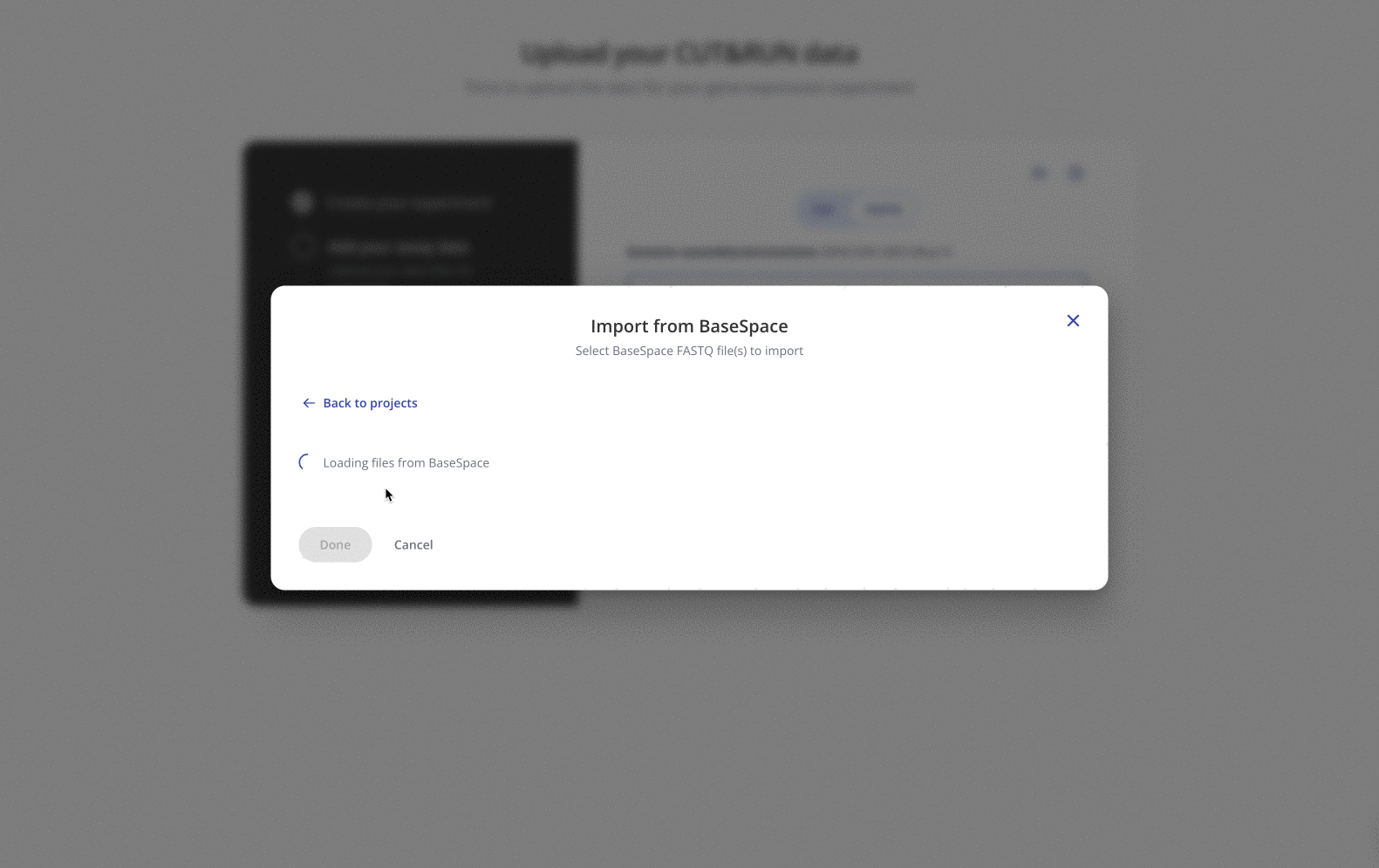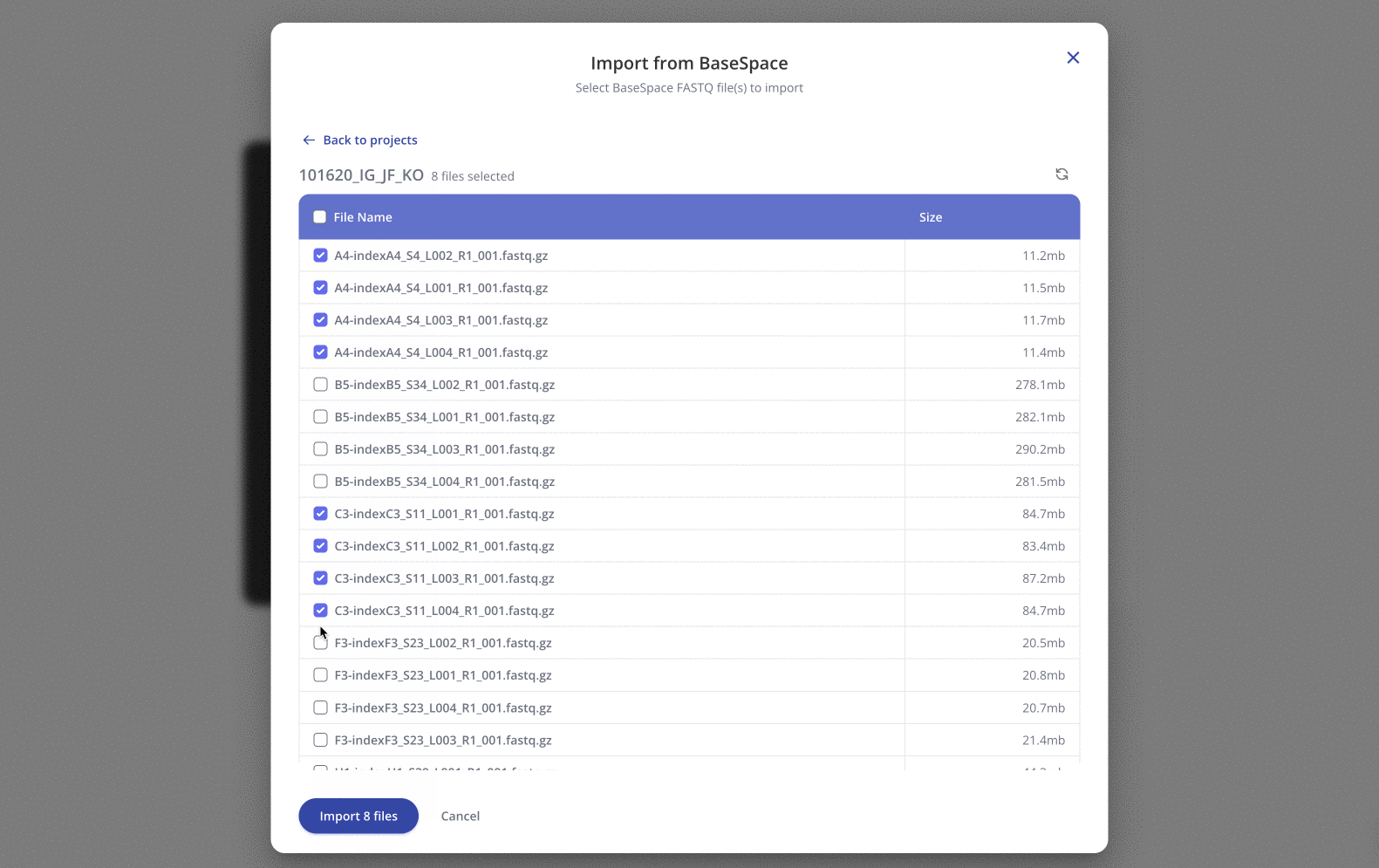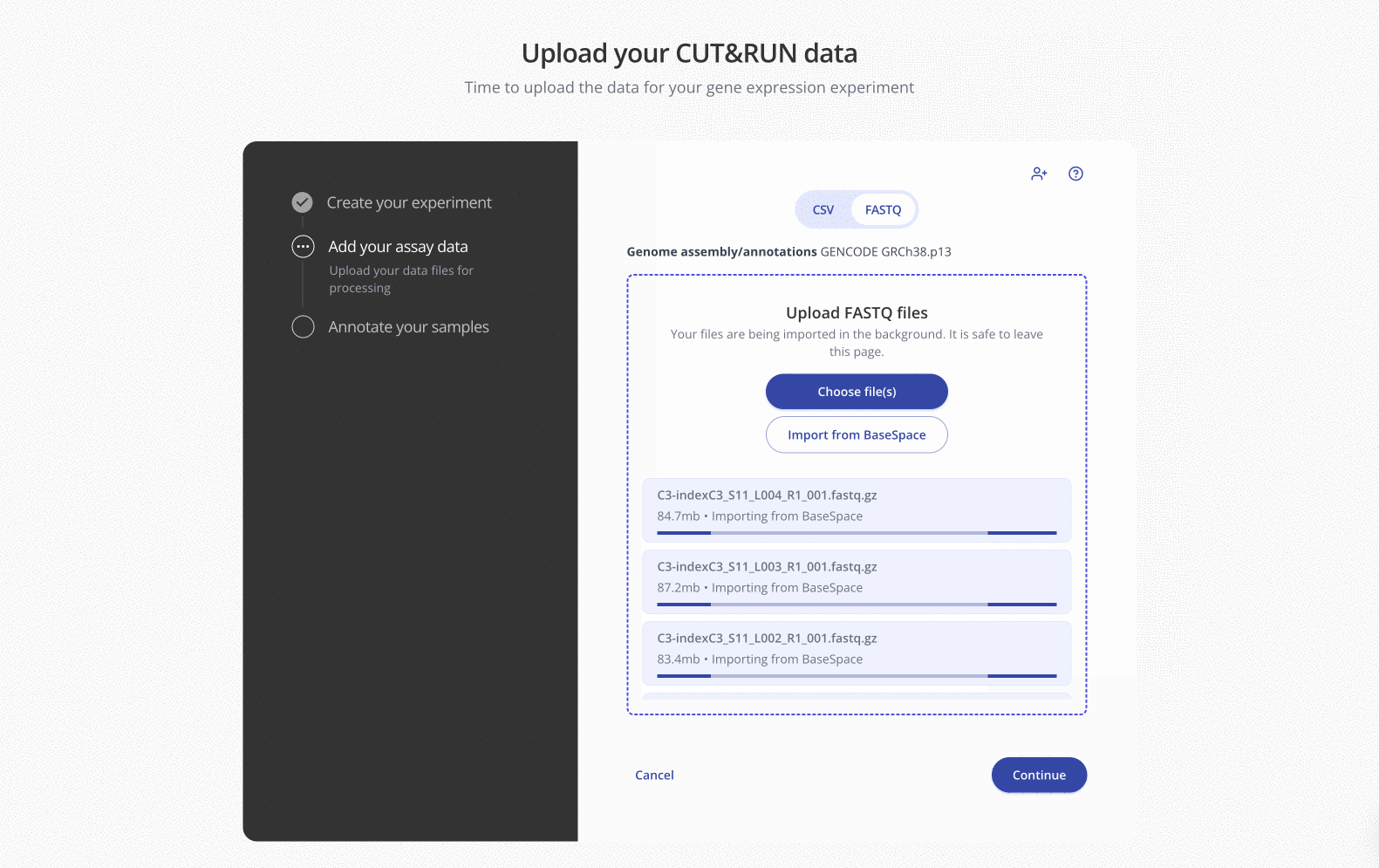
In the pop-up window, simply navigate into the appropriate BaseSpace project and select the FASTQs you'd like to import.
