Easily upload preprocessed single-cell RNA-seq Seurat objects into Pluto for seamless downstream analysis and visualization
Uploading Seurat objects into Pluto enables you to leverage preprocessed single-cell RNA-seq data for in-depth downstream analysis. Whether you’ve already performed analysis externally or are pulling data from public sources, you can now skip the initial steps of data processing and start your exploration immediately within Pluto. This functionality is especially useful for accelerating your research, as it allows you to take your existing Seurat datasets and dive directly into the analysis, uncovering insights without the need to reprocess or cluster the data again. By easily importing Seurat objects, you gain the flexibility to address your specific scientific questions and explore data from a variety of sources, all within the Pluto platform.What is a Seurat object?
A Seurat object is a data structure used in the Seurat R package for single-cell RNA sequencing (RNA-seq) analysis. It holds various types of data related to your single-cell experiment, including gene expression values, metadata, and results from dimensionality reduction and clustering. Seurat objects allow you to store all the components of your analysis in one container for easy access and manipulation.
Seurat object structure
A Seurat object consists of several key components:
-
Assays: These are different types of data stored in the Seurat object, such as gene expression data. Each assay can have multiple slots, typically one for the raw data and one for processed data (e.g., normalized or scaled expression data).
-
Metadata: This contains information about the cells in your experiment. It might include cell type information, experimental conditions, sample labels, and more. This metadata is crucial for grouping, filtering, and analyzing your data in a meaningful way.
-
Dimensionality reduction coordinates: These are the results of techniques like PCA, UMAP, or tSNE, which reduce the high-dimensional data (e.g., gene expression values) to fewer dimensions for easier visualization and interpretation. These coordinates are stored in the Seurat object as part of the object’s reductions slot.
-
Clusters: The Seurat object may also contain clustering results. These are the groupings of cells that have been assigned based on their similarities. Clusters may or may not be annotated with cell type identities.
What is an RDS file?
The Seurat object is typically saved as an RDS file in R, which allows for efficient storage and easy sharing of the object. This format preserves the structure and data of the Seurat object, including metadata, assays, and reductions, making it easy to reload and continue working with the object later.
Preparing your Seurat object for upload to Pluto
Before uploading your Seurat object into Pluto, there are a few important steps to ensure your object is in the correct format. The Seurat object must be preprocessed and contain dimensionality reduction results, clustering results, and sample-level data. Let’s go through each of these requirements.
Dimensionality reduction results
The Seurat object should contain results from dimensionality reduction methods such as PCA, UMAP, and / or tSNE. These results will be stored in the Seurat object under the reductions slot.
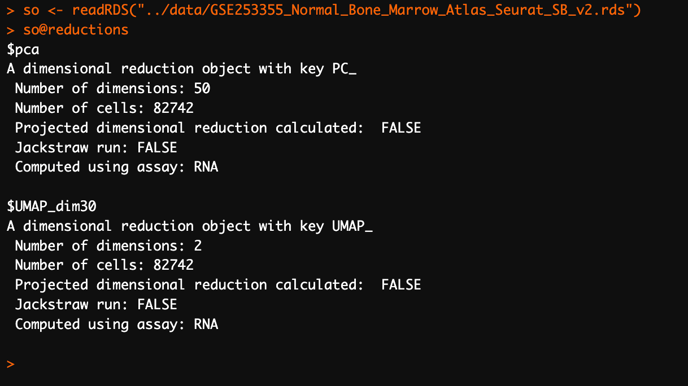
In the above example, the Seurat object
so has two dimensional reductions: PCA (pca) and UMAP (UMAP_dim30).Clustering results
The Seurat object should contain clustering results, either as unannotated clusters (e.g., numeric cluster labels) or annotated clusters (e.g., cell type annotations). The clusters will be used for downstream analysis and visualization in Pluto.
If your clusters are annotated with cell type information (e.g., cluster 1 is "T cells", cluster 2 is "B cells"), Pluto will display these annotations once the object is uploaded. If your clusters are unannotated, you can still upload the object and use Pluto to assign cell type identities to the clusters based on your analysis.
To check clustering results in your Seurat object, load your Seurat object into R and check the Seurat objectmeta.data for columns that corresponds to clusters / cell type annotations: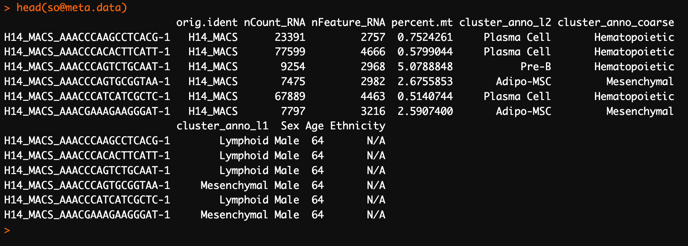
In the above example, the Seurat object
so has three columns that correspond to annotated clusters: cluster_anno_l2, cluster_anno_coarse, and cluster_anno_l1Sample-level data
Your Seurat object should include sample-level data within its metadata, as this is essential for properly grouping and identifying samples during analysis. Unlike cell-level data, which represents measurements or gene expression values for individual cells, sample-level data consists of aggregated or summary information across all cells within a given sample or condition. Sample-level data might include information such as:
- Sample names
- Experimental conditions
- Batch information
Sample-level data is stored in the meta.data slot of the Seurat object.
To check if sample-level data has been incorporated into your Seurat object, load your Seurat object into R and check the Seurat object meta.data for columns that correspond to sample-level data:

In the above example, the following columns correspond to sample-level data: orig.ident, Sex, Age, and Ethnicity.
Uploading your Seurat object to Pluto
Once your Seurat object meets all the requirements, uploading it to Pluto is simple.
From the Experiment wizard, under "Add your assay data", select the "RDS" button and drag-and-drop in your Seurat object - saved as a .RDS file - or search for your Seurat object locally by clicking "Choose file".

Once you are done uploading your Seurat object, confirm that you are ready to move on. After a Seurat object has been uploaded to Pluto, you will no longer be able to modify it. This is to ensure the integrity of the data during the analysis process. Any changes you want to make (such as re-running preprocessing or clustering) should be done prior to uploading the Seurat object into Pluto.

After successfully uploading your Seurat object, it will be seamlessly integrated into your Pluto workspace. Once the Seurat object is added to your workspace, you will receive an email notification and can start analyzing your data in Pluto. Whether you’re visualizing clusters, identifying cell types, or exploring other aspects of your dataset, Pluto allows you to take full advantage of your preprocessed Seurat object for further exploration and discovery.

Need help?
If you encounter any issues or need assistance preparing your Seurat object for upload, please reach out to our support team at support@pluto.bio. We’re here to help you make the most of Pluto’s features and ensure a smooth analysis experience.
