Use the STRING database to annotate protein-protein interactions in your experimental data
In Pluto, you can use the Protein-protein interaction analysis module to annotate proteins using the well-known STRING database.
1. Add a new analysis and select protein-protein interaction analysis under the Pathway category.
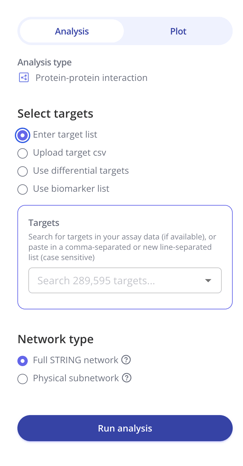
2. Select how you'd like to provide genes/proteins to the protein-protein interaction analysis. You can manually enter a list of gene/protein symbols, upload a CSV file, use a DEG table as input, or use one of your saved lists. Click Run analysis to query the STRING database and generate a network.
3. Customize your network graph appearance and toggle on/off different kinds of associations, which are shown as the different colored lines connecting each node (protein).
