Visit the Pipeline tab to generate a dynamic methods summary for your experiment
For experiments containing raw assay data (e.g. FASTQ files) in Pluto, you can run a variety of pipelines, which transform raw data into a processed/tabular data format. Pipelines available in Pluto include:
-
Bulk RNA-seq
-
scRNA-seq
-
ChIP-seq
-
CUT&RUN
-
ATAC-seq
After uploading raw data and sample annotations to Pluto, you'll select parameters to configure the pipeline run for your experiment. These parameters will vary based on the type of experiment.
Recalling pipeline parameters in Pluto
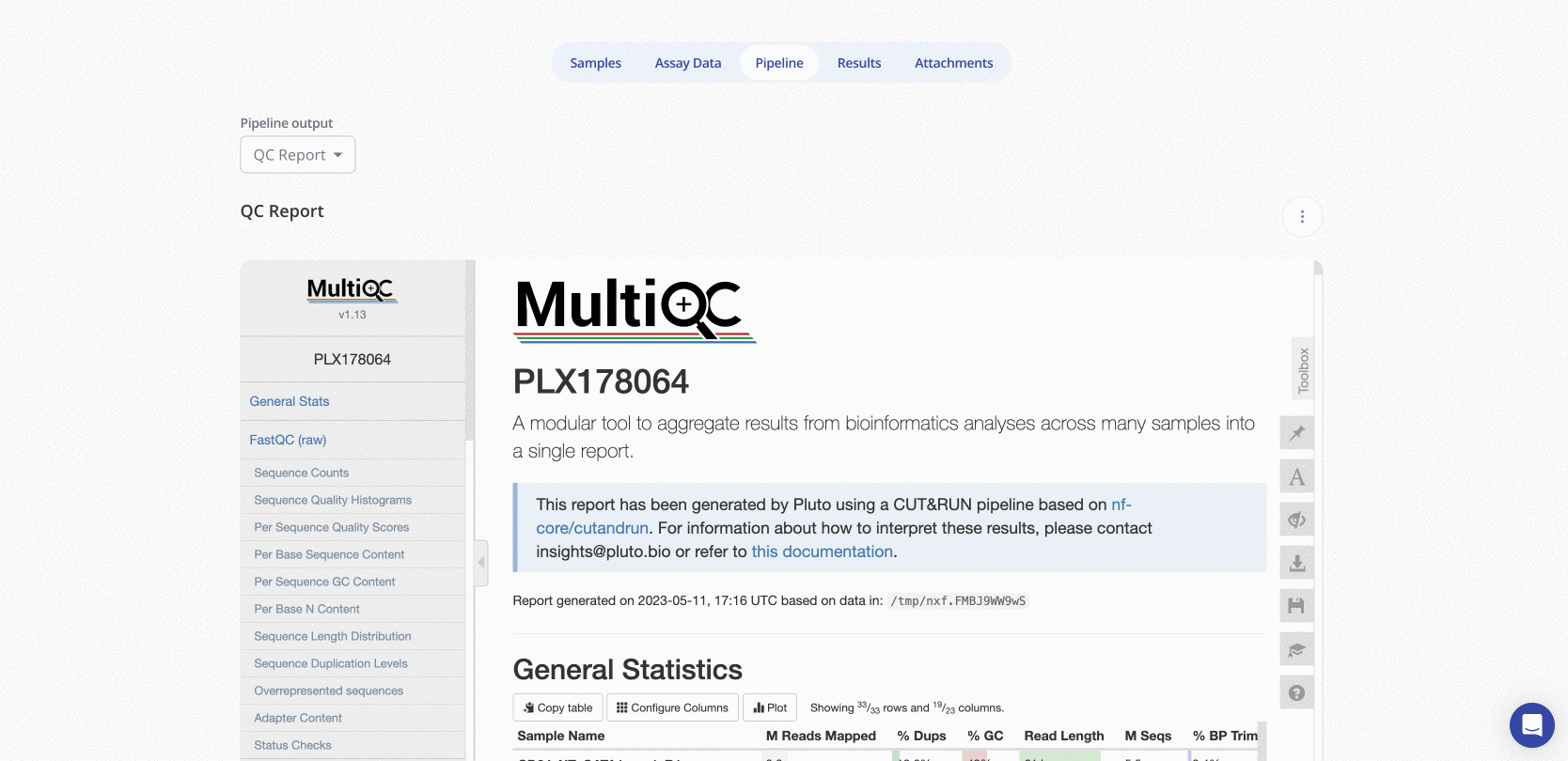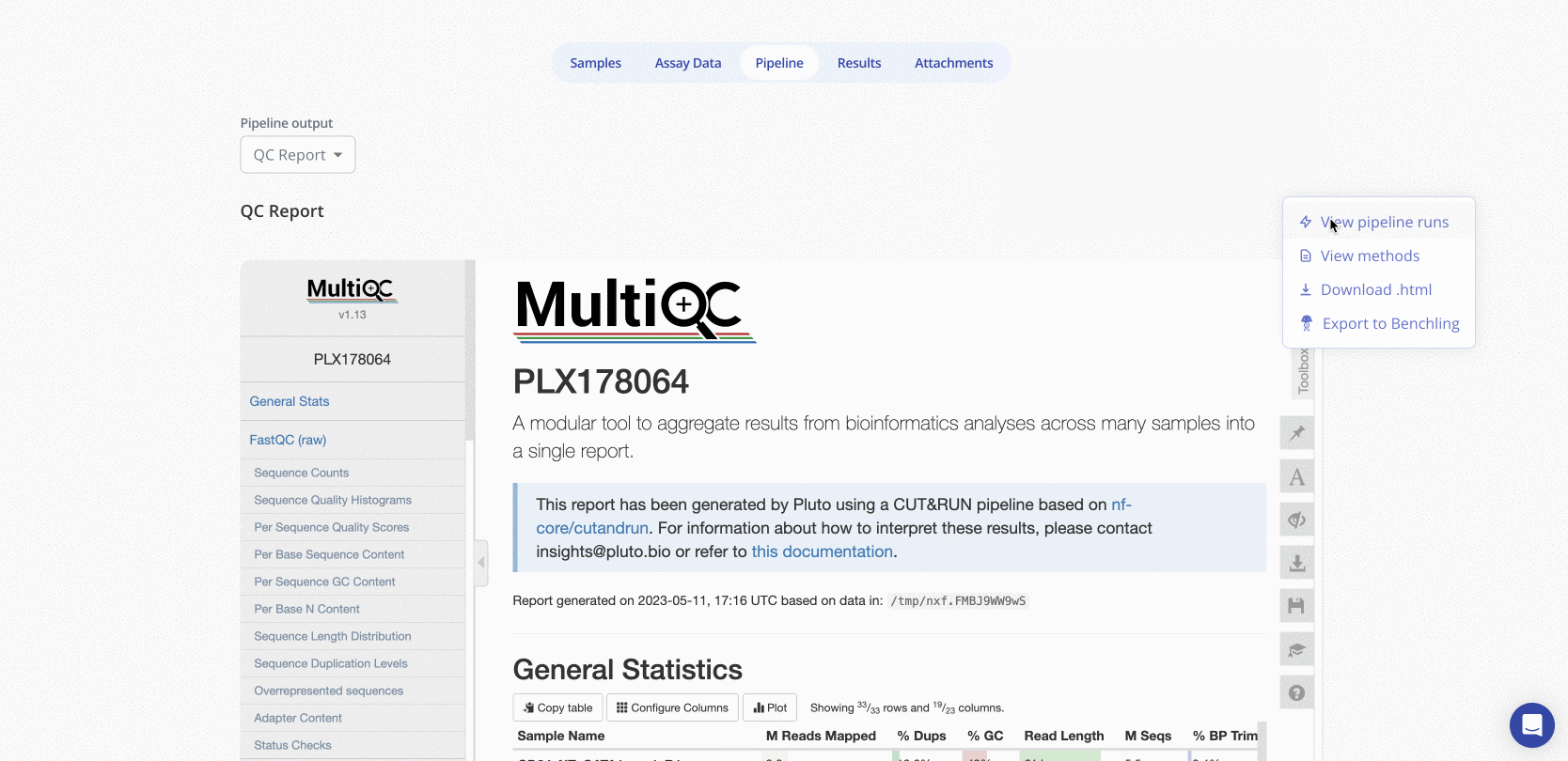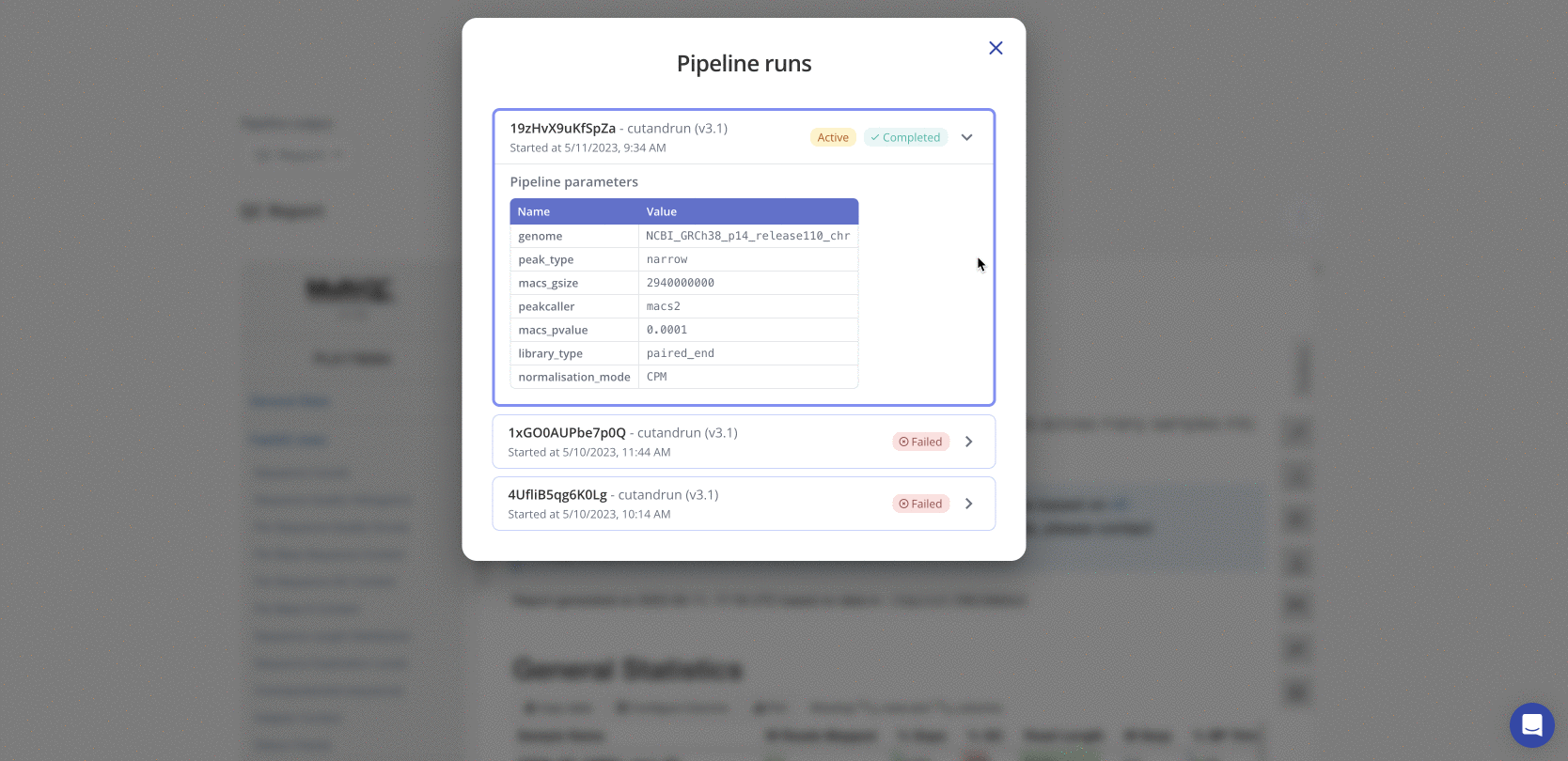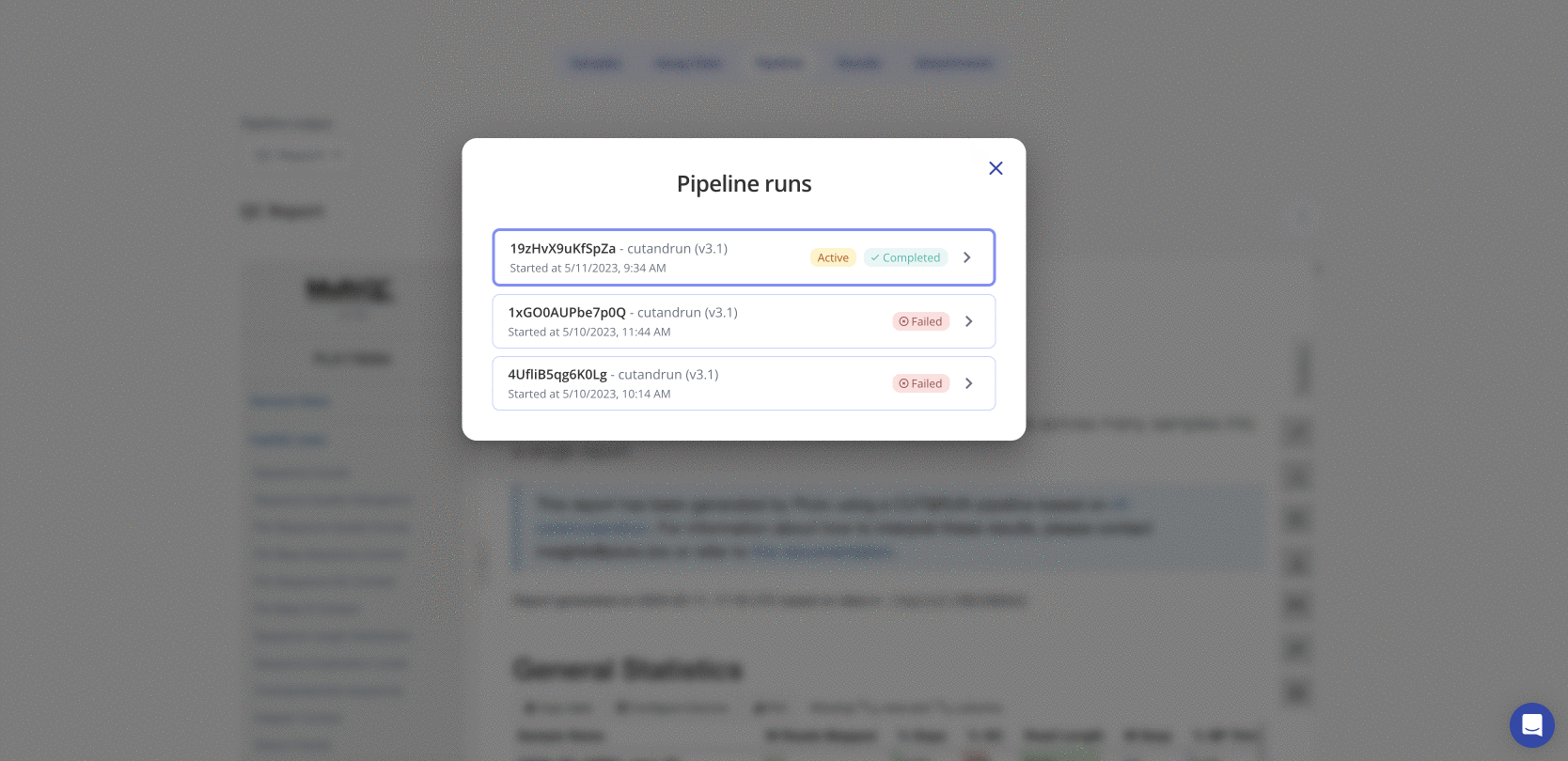
Navigate to the Pipeline tab on the experiment page in Pluto and select "View pipeline runs" to view all prior pipeline runs for that experiment. If multiple pipeline runs exist, the Active pipeline indicates the pipeline for which final data is shown in Pluto. Click on any pipeline run to expand the list of user-selected parameters that were used for that run.
Viewing the pipeline methods section in Pluto
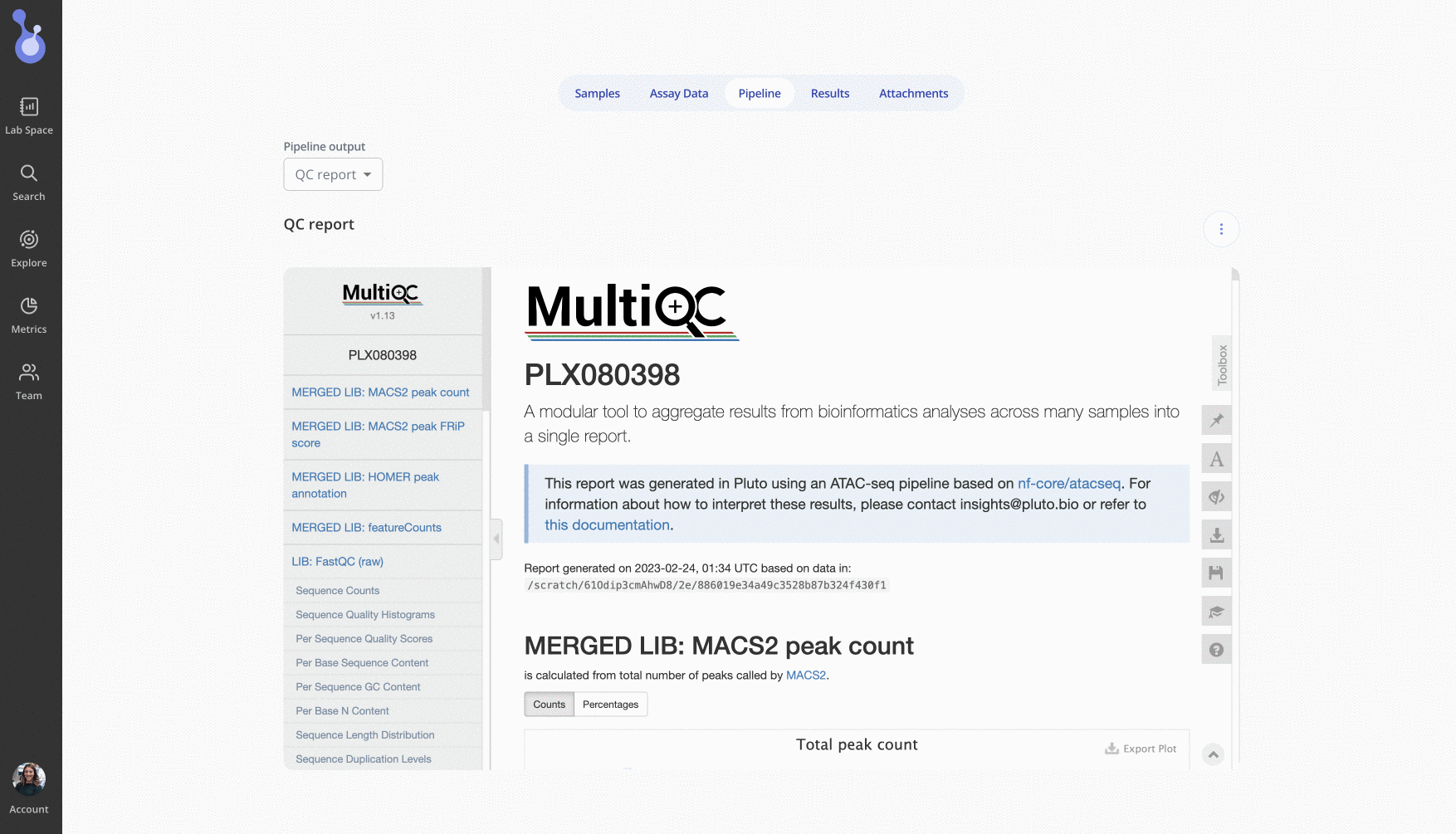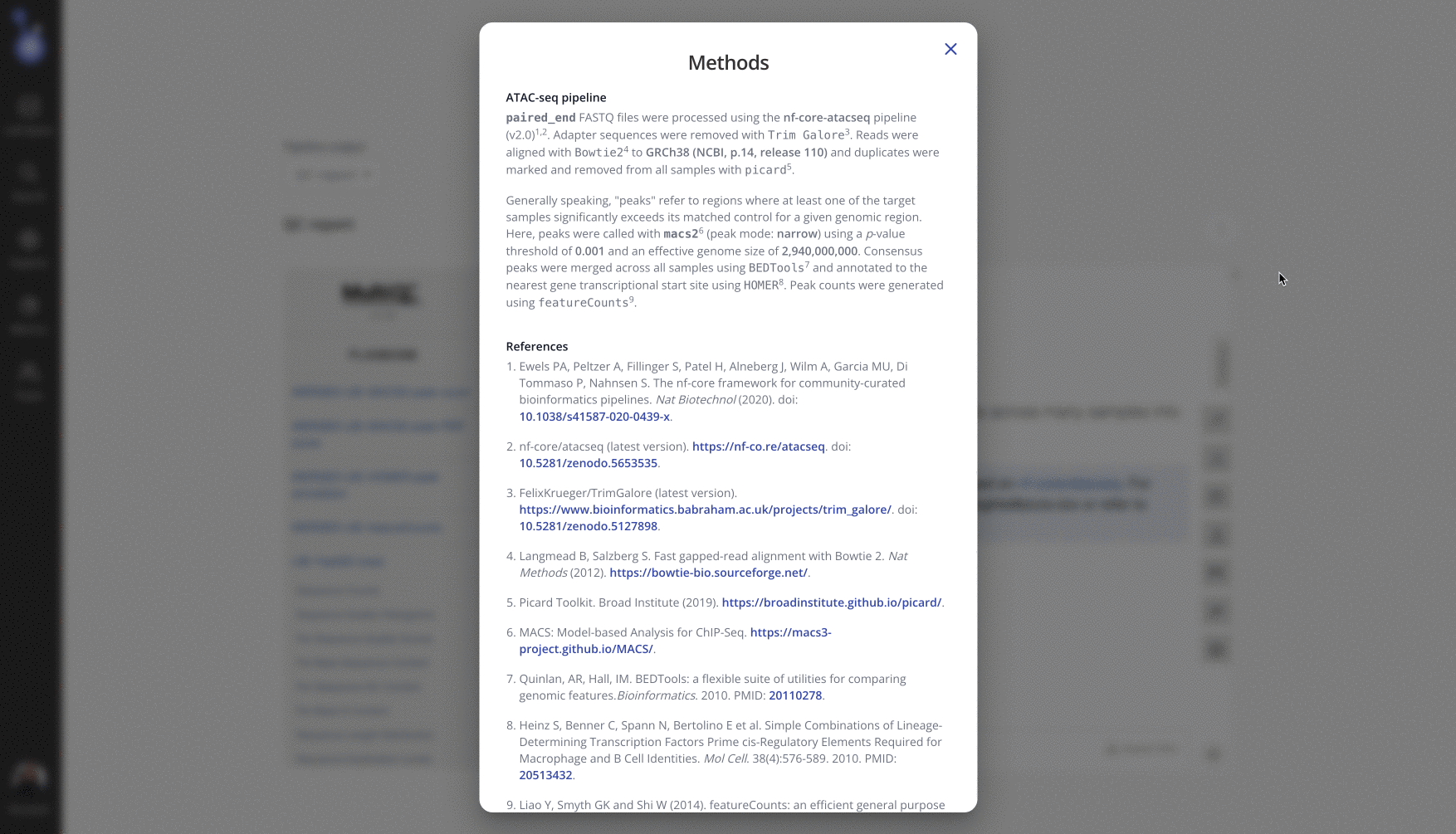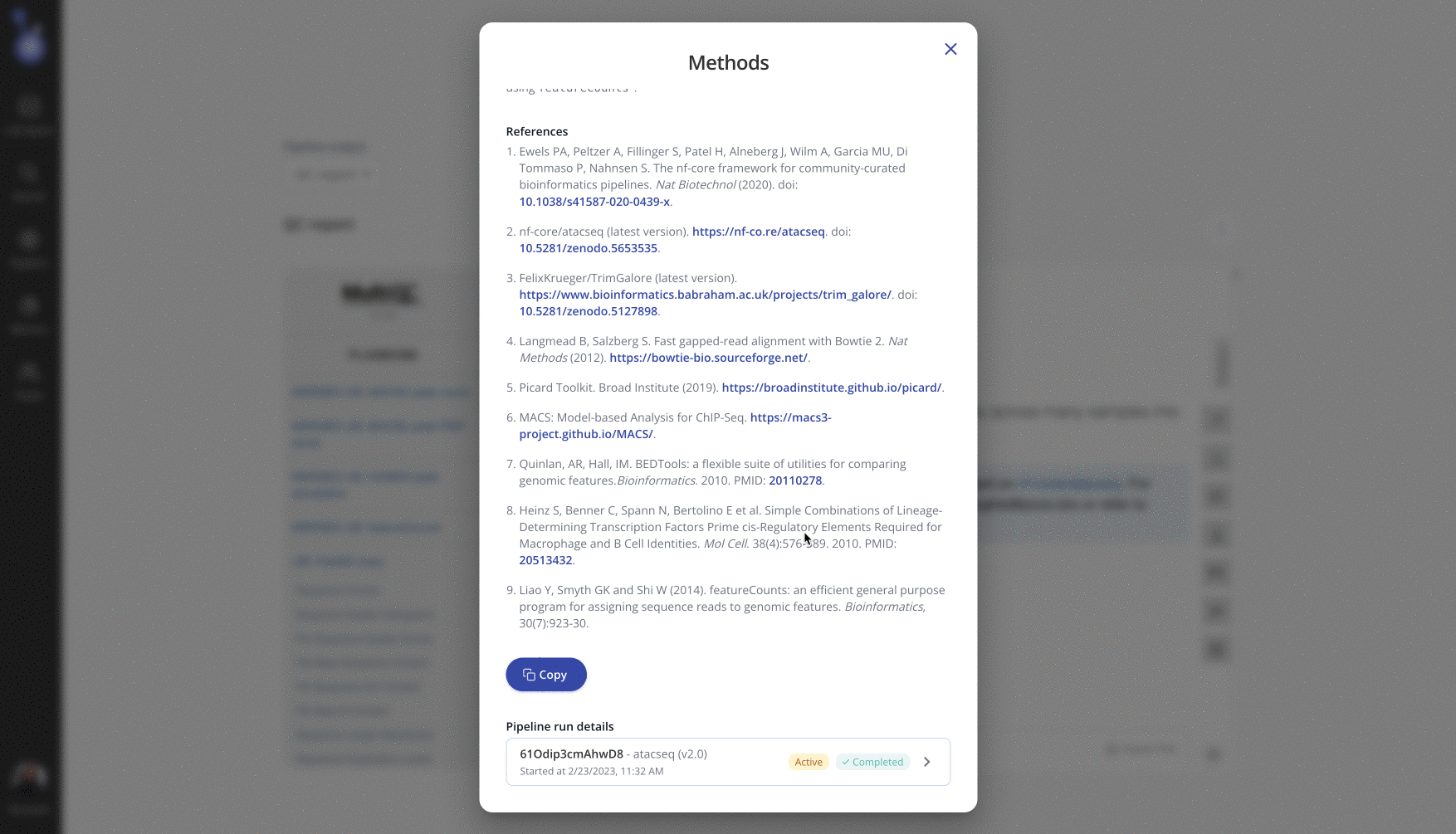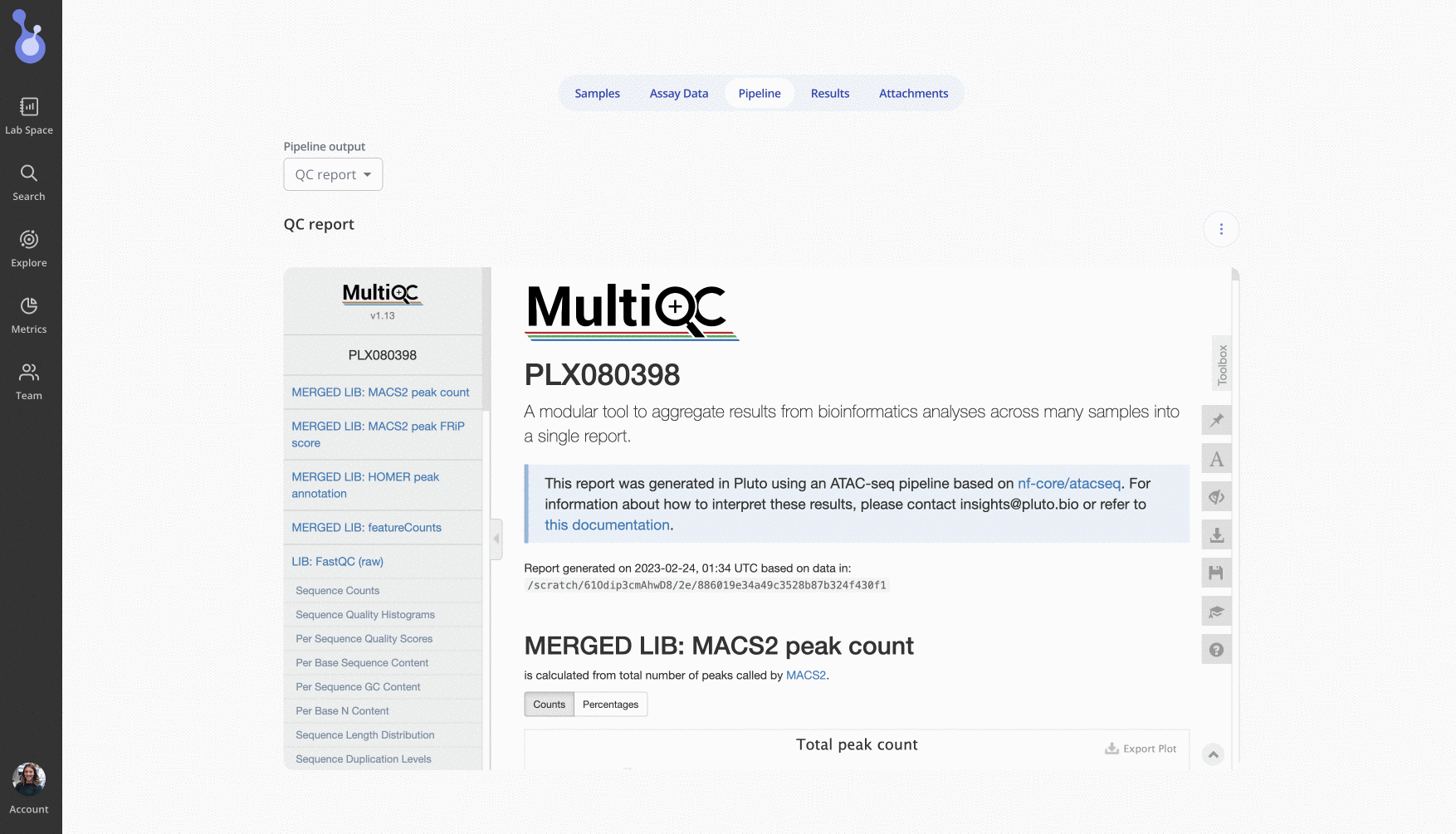
From the Pipeline tab on the experiment page in Pluto, select the "..." menu > "View methods". This dynamic methods section contains details specific to the user-selected parameters used for this experiment, as well as references to any relevant software tools. Use the "Copy" button to copy the dynamic methods section to paste into your manuscript.
You can also download pipeline intermediate files from the Pipeline tab.
